| Instructions | Output format | Data | Software |
ApoplastP: prediction of effectors and plant proteins in the apoplast using machine learning
Introduction
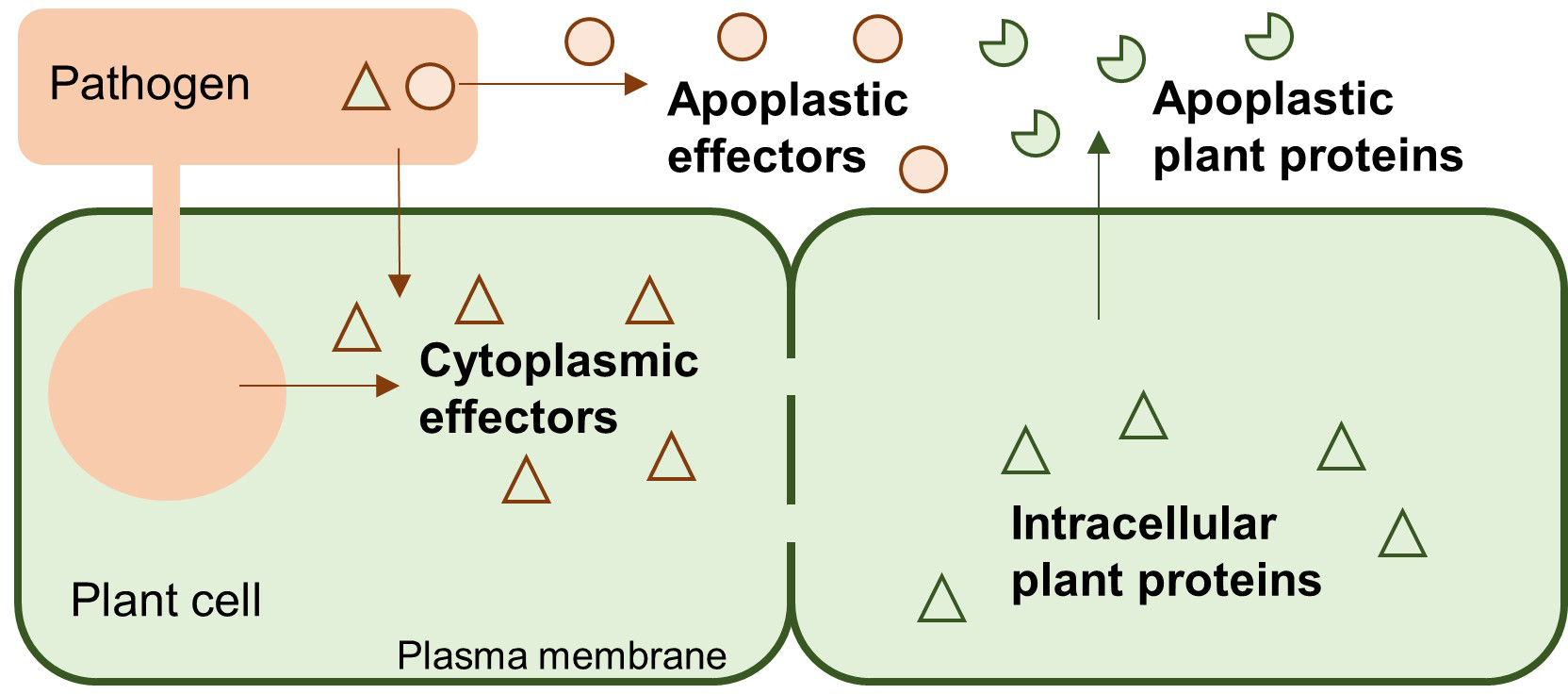
ApoplastP is a machine learning method for predicting localization of proteins to the plant apoplast. ApoplastP can distinguish non-apoplastic proteins from apoplastic proteins for both plant proteins and pathogen proteins. In particular, ApoplastP can predict if an effector localizes to the plant apoplast.
Submission
To run ApoplastP, please submit your proteins of interest below. For online submission, the maximum number of protein sequences that can be submitted is 4000. If you want to run ApoplastP on your local machine, you can download the current version here.
Note that predictions for protein sequences with excessive unknown bases (X) are unreliable.
Citation
Contact
For comments, suggestions and reporting bugs please contact jana.sperschneider@csiro.au
